What is this?
This site provides BMRB NMR-STAR data derived from NMR spectroscopy in XML, RDF and JSON formats. BMRB/XML, BMRB/RDF and BMRB/JSON are names of data collections, respectively.
About BMRB
BMRBj (Biological Magnetic Resonance Data Bank Japan) and Biological Magnetic Resonance Data Bank (BMRB in UConn) are a common repository for experimental and derived data gathered from nuclear magnetic resonance (NMR) spectroscopic studies of biological molecules. As a member of the Worldwide Protein Data Bank (wwPDB), the BMRB maintains archives of quantitative NMR spectral parameters (e.g. assigned chemical shifts, J-coupling constants, etc.), derived data (e.g. relaxation parameters, kinetics parameters, NMR restraints used to determining the structures), time-domain spectral data (FID) and a metabolite NMR spectral database. The BMRB and BMRBj provide data deposition, validation and visualization tools in collaboration with other wwPDB members (RCSB-PDB, PDBj, PDBe).
Representing an alliance of BMRBs (USA at University of Connecticut, Japan at Osaka University and Europe at University of Florence), BMRBj provides the NMR-STAR data in web standard formats, XML and RDF to enhance the interoperability of the BMRB archival data.
BMRB/XML, BMRB/RDF, and BMRB/JSON
Schema of the BMRB/XML (BMRB/XML Schema) is translated from the current NMR-STAR v3 Dictionary and make it easy for scientists familiar with NMR-STAR files to comprehend the contents in the XML format. BMRB/XML accommodates a rich information content in a single file including entry citations, experimental parameters, assigned chemical shifts, links to external databases, atom coordinates of the biomolecules, NMR restraints, assigned peak lists, etc.
The BMRB/RDF files are generated by direct translation from the corresponding BMRB/XML data files. Along with the BMRB/RDF, we provide its ontology of the BMRB/RDF (BMRB/OWL) in Web Ontology Language (OWL). The RDF/RDFS/OWL Semantic Web flamework constitute a standard model for data exchange on the Web, and make it possible to query across diverse data sources using SPARQL.
NMR-STAR format family
| Format | NMR-STAR | NMR-STAR (RDBMS) | BMRB/XML | BMRB/RDF | BMRB/JSON |
|---|---|---|---|---|---|
| Ontology | Canonical NMR-STAR v3 Dictionary Extended dictionary for BMRB/XML |
Database DDL DDL based on BMRB/XML Schema DDL compatible with NMR-STAR Dic. |
BMRB/XML Schema | BMRB/OWL | BMRB/JSON Schema |
| Validation | ADIT-NMR, Manual annotation using in-house validator | in-house DB loader | Automated remediation by BMRBxTool Full XML Schema validation by Apache Xerces |
RDF validation by BMRBoTool with Redland Raptor | Automated remediation by BMRBxTool JSON Schema validation by daveclayton/json-schema-validator |
| Data type check | ADIT-NMR, Manual annotation using in-house validator (string based) |
Database schema (DDL based) |
XML Schema data types (ID, string, integer, real, date, enumeration) Metadata check referring to external DBs (PubMed, Taxonomy, PDB, Ligand Expo, etc.) |
Inheriting all XML Schema data types | Inheriting all XML Schema data types |
| Type of storage | Text files (.str) | RDBMS | XML files (.xml), XML Database | RDF files (.rdf, .nt), RDF triplestore | JSON files (.json), NoSQL Database |
| Standardization | |||||
| Serialization | |||||
| Parser libraries | starlibj, starlib2, SANS, and PyNMRSTAR | JDBC, ODBC drivers and various language bindings | DOM, SAX, StAX and JAXB libraries and various language bindings | RDF parsing libralies, RDF triplestore | ECMAScript or various JSON parser libraries and their language bindings |
| Query languages | Not standardized, but you can do with either starlibj, starlib2 or SANS | SQL | XPath, XSLT, XQuery | SPARQL | ECMAScript, JSONiq, XQeury 3.1 |
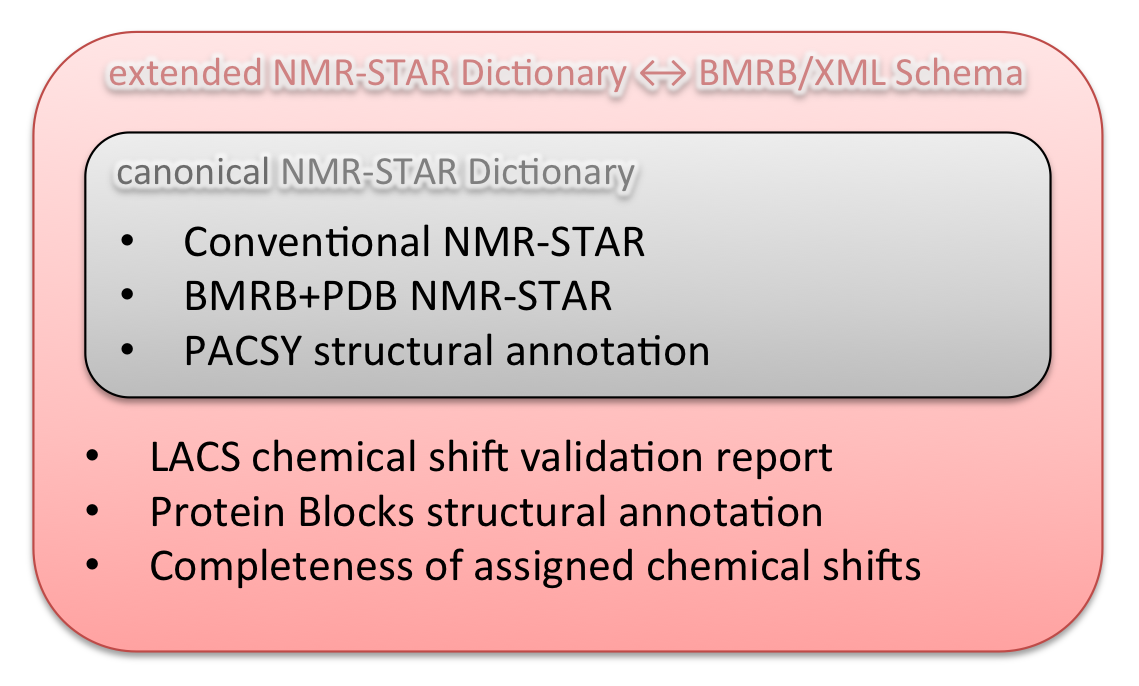
| Information content | Canonical repositories: Conventional BMRB+PDB Metabolomics Extended repositories: LACS validation report Protein Blocks annotation CS Completeness |
Conventional + "BMRB+PDB" + PACSY and Metabolomics link to PACSY structural annotation BMRB RDB Snapshot an alternative RDB distribution service compatible with BMRB/XML "noatom" version. |
"complete" and "noatom" "complete" accommodates the next repositories: Conventional, BMRB+PDB, PACSY, LACS, Protein Blocks, CS completeness. "noatom" omits coordinate, restraints and peak lists from the "complete". |
RDF/XML or N-Triples Both accomodate information content of "noatom" version of BMRB/XML. |
"noatom" |
Comparison of information contents
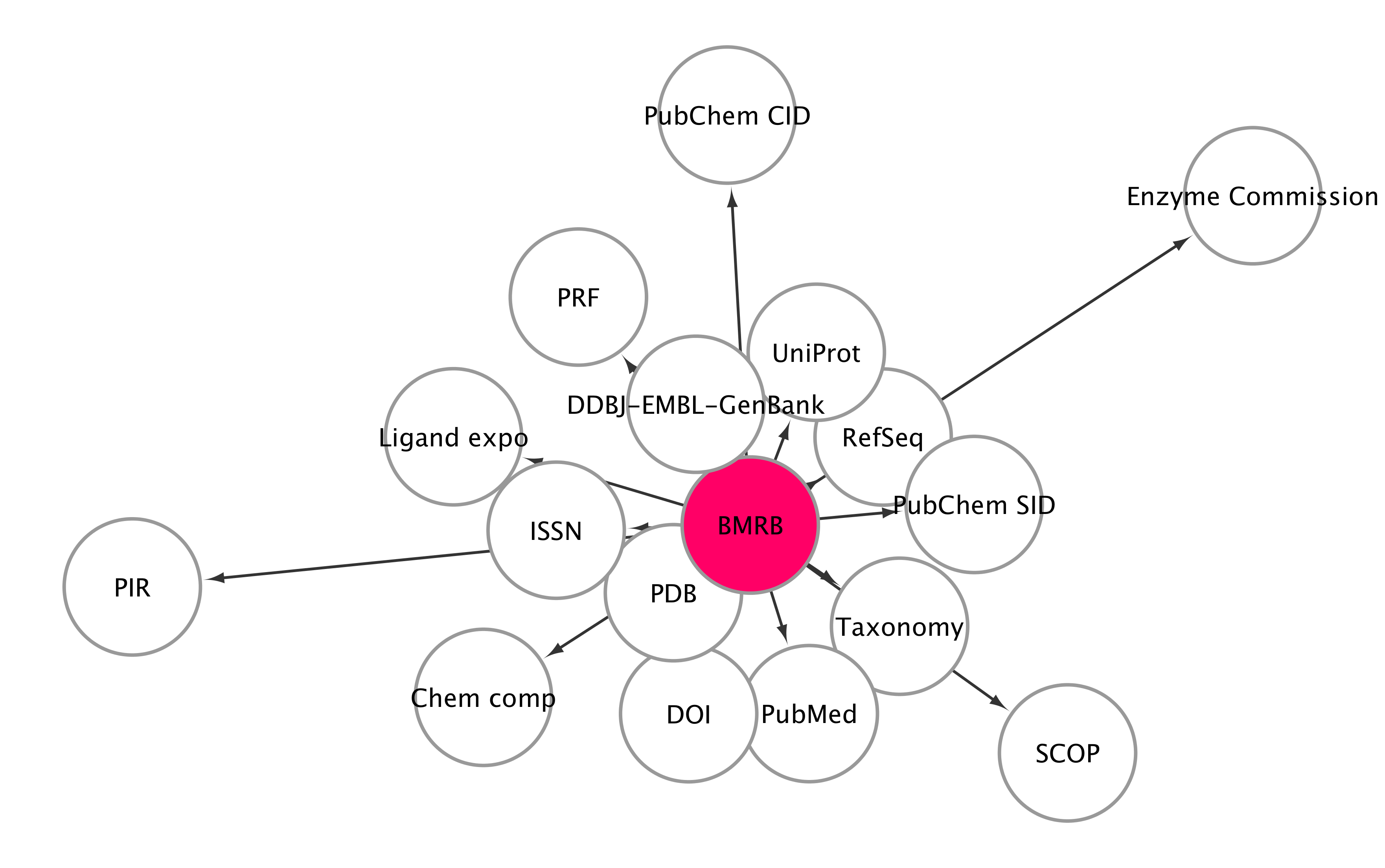
Linked data with BMRB
References
Bekker GJ, Yokochi M, Suzuki H, Ikegawa Y, Iwata T, Kudou T, Yura K, Fujiwara T, Kawabata T, Kurisu G, Protein Data Bank Japan: Celebrating our 20th anniversary during a global pandemic as the Asian hub of three dimensional macromolecular structural data, Protein Science, 31(1), 173-186 (2022)
Kinjo AR, Bekker GJ, Wako H, Endo S, Tsuchiya Y, Sato H, Nishi H, Kinoshita K, Suzuki H, Kawabata T, Yokochi M, Iwata T, Kobayashi N, Fujiwara T, Kurisu G, Nakamura H, New tools and functions in data-out activities at Protein Data Bank Japan (PDBj), Protein Science, 27(1), 95-102 (2018)
Yokochi M, Kobayashi N, Ulrich EL, Kinjo AR, Iwata T, Ioannidis YE, Linvy M, Markley JL, Nakamura H, Kojima C, Fujiwara T, Publication of nuclear magnetic resonance experimental data with semantic web technology and the application thereof to biomedical research of proteins, J. Biomed. Semantics, 7(1), 16 (2016)
Ulrich EL, Akutsu H, Doreleijers JF, Hrano Y, Ioannidis YE, Lin J, Linvy M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Wenger RK, Yao H, Markley JL, BioMagResBank, Nucleic Acids Research, 36, D402-D408 (2008)
Ulrich EL, Baskaran K, Dashti H, Ioannidis YE, Livny M, Romero PR, Maziuk D, Wedell JR, Yao H, Eghbalnia HR, Hoch JC, Markley JL, NMR-STAR: comprehensive ontology for representing, archiving and exchanging data from nuclear magnetic resonance spectroscopic experiments, J. biomol. NMR, ePub ahead, 1-5 (2018)
Kawashima S, Katayama T, Hatanaka H, Kushida T, Takagi T, NBDC RDF portal: a comprehensive repository for semantic data in life sciences, Database, 2018, bay123, 1-11 (2018)



